ProbeFeatureSelection#
ProbeFeatureSelection() generates one or more random variables based on the
user-selected parameters. Next, the transformer derives the feature importance for each variable
and probe feature. Finally, it eliminates the features that have a lower feature importance
score than the probe feature(s).
In the case of there being more than one probe feature, the average feature importance score of all the probe features is used.
In summary, this is how ProbeFeatureSelection() selects features:
Create 1 or more random features
Train a machine learning model with all features including the random ones
Derive feature importance for all features
Take the average importance of the random features (only if more than 1 random feature were used)
Select features whose importance is greater than the importance of the random variables (step 4)
One of the primary goals of feature selection is to remove noise from the dataset. A randomly generated variable, i.e., probe feature, inherently possesses a high level of noise. Consequently, any variable that demonstrates less importance than a probe feature is assumed to be noise and can be discarded from the dataset.
When initiating the ProbeFeatureSelection() class, the user has the option of selecting
which distribution is to be assumed to create the probe feature(s) and the number of
probe features to be created. The possible distributions are ‘normal’, ‘binary’, ‘uniform’,
or ‘all’. ‘all’ creates 1 or more probe features comprised of each distribution type,
i.e., normal, binomial, and uniform.
Example#
Let’s see how to use this transformer to select variables from UC Irvine’s Breast Cancer Wisconsin (Diagnostic) dataset, which can be found here. We will use Scikit-learn to load the dataset. This dataset concerns breast cancer diagnoses. The target variable is binary, i.e., malignant or benign.
The data is solely comprised of numerical data.
Let’s import the required libraries and classes:
import pandas as pd
from sklearn.datasets import load_breast_cancer
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split
from feature_engine.selection import ProbeFeatureSelection
Let’s now load the cancer diagnostic data:
cancer_X, cancer_y = load_breast_cancer(return_X_y=True, as_frame=True)
Let’s check the shape of cancer_X:
print(cancer_X.shape)
We see that the dataset is comprised of 569 observations and 30 features:
(569, 30)
Let’s now split the data into train and test sets:
# separate train and test sets
X_train, X_test, y_train, y_test = train_test_split(
cancer_X,
cancer_y,
test_size=0.2,
random_state=3
)
X_train.shape, X_test.shape
We see the size of the datasets below. Note that there are 30 features in both the training and test sets.
((455, 30), (114, 30))
Now, we set up ProbeFeatureSelection().
We will pass RandomForestClassifier() as the estimator. We will use precision
as the scoring parameter and 5 as cv parameter, both parameters to be
used in the cross validation.
In this example, we will introduce just 1 random feature with a normal distribution. Thus,
we pass 1 for the n_probes parameter and normal as the distribution.
sel = ProbeFeatureSelection(
estimator=RandomForestClassifier(),
variables=None,
scoring="precision",
n_probes=1,
distribution="normal",
cv=5,
random_state=150,
confirm_variables=False
)
sel.fit(X_train, y_train)
With fit(), the transformer:
creates
n_probesnumber of probe features using provided distribution(s)uses cross-validation to fit the provided estimator
calculates the feature importance score for each variable, including probe features
if there are multiple probe features, the transformer calculates the average importance score
identifies features to drop because their importance scores are less than that of the probe feature(s)
In the attribute probe_features, we find the pseudo-randomly generated variable(s):
sel.probe_features_.head()
gaussian_probe_0
0 -0.694150
1 1.171840
2 1.074892
3 1.698733
4 0.498702
We can go ahead and display a histogram of the probe feature:
sel.probe_features_.hist(bins=30)
As we can see, it shows a normal distribution:
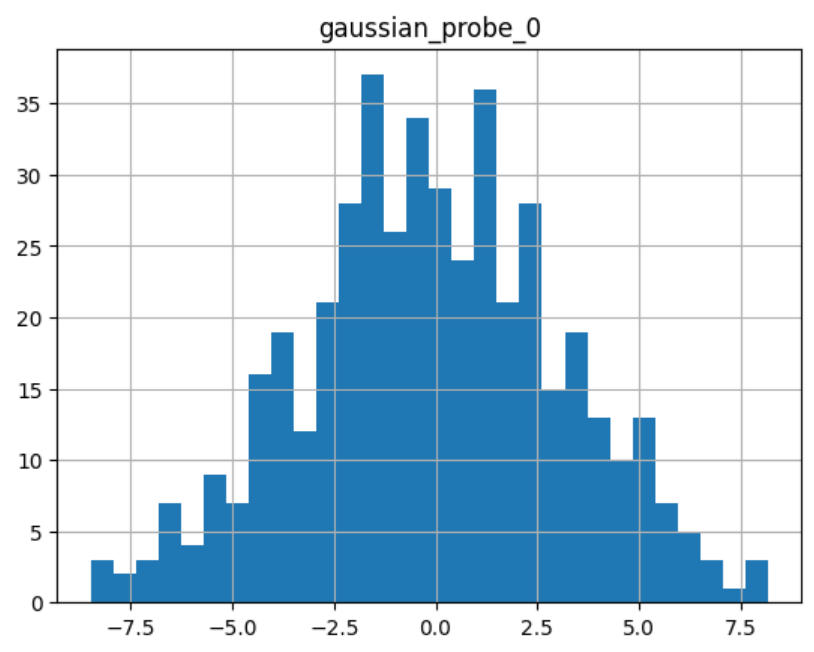
The attribute feature_importances_ shows each variable’s feature importance:
sel.feature_importances_.head()
These are the first 5 features:
mean radius 0.058463
mean texture 0.011953
mean perimeter 0.069516
mean area 0.050947
mean smoothness 0.004974
At the end of the series, we see the importance of the probe feature:
sel.feature_importances_.tail()
These are the importance of the last 5 features including the probe:
worst concavity 0.037844
worst concave points 0.102769
worst symmetry 0.011587
worst fractal dimension 0.007456
gaussian_probe_0 0.003783
dtype: float64
In the attribute features_to_drop_, we find the variables that were not selected:
sel.features_to_drop_
These are the variables that will be removed from the dataframe:
['mean symmetry',
'mean fractal dimension',
'texture error',
'smoothness error',
'concave points error',
'fractal dimension error']
We see that the features_to_drop_ have feature importance scores that are less
than the probe feature’s score:
sel.feature_importances_.loc[sel.features_to_drop_+["gaussian_probe_0"]]
The previous command returns the following output:
mean symmetry 0.003698
mean fractal dimension 0.003455
texture error 0.003595
smoothness error 0.003333
concave points error 0.003548
fractal dimension error 0.003576
gaussian_probe_0 0.003783
With transform(), we can go ahead and drop the six features with feature importance score
less than gaussian_probe_0 variable:
Xtr = sel.transform(X_test)
Xtr.shape
The final shape of the data after removing the features:
(114, 24)
And, finally, we can also obtain the names of the features in the final transformed dataset:
sel.get_feature_names_out()
['mean radius',
'mean texture',
'mean perimeter',
'mean area',
'mean smoothness',
'mean compactness',
'mean concavity',
'mean concave points',
'radius error',
'perimeter error',
'area error',
'compactness error',
'concavity error',
'symmetry error',
'worst radius',
'worst texture',
'worst perimeter',
'worst area',
'worst smoothness',
'worst compactness',
'worst concavity',
'worst concave points',
'worst symmetry',
'worst fractal dimension']
For compatibility with Scikit-learn selection transformers, ProbeFeatureSelection()
also supports the method get_support():
sel.get_support()
which returns the following output:
[True, True, True, True, True, True, True, True, False, False, True, False, True,
True, False, True, True, False, True, False, True, True, True, True, True, True,
True, True, True, True]
Using several probe features#
Let’s now repeat the selection process, but using more than 1 probe feature.
sel = ProbeFeatureSelection(
estimator=RandomForestClassifier(),
variables=None,
scoring="precision",
n_probes=3,
distribution="all",
cv=5,
random_state=150,
confirm_variables=False
)
sel.fit(X_train, y_train)
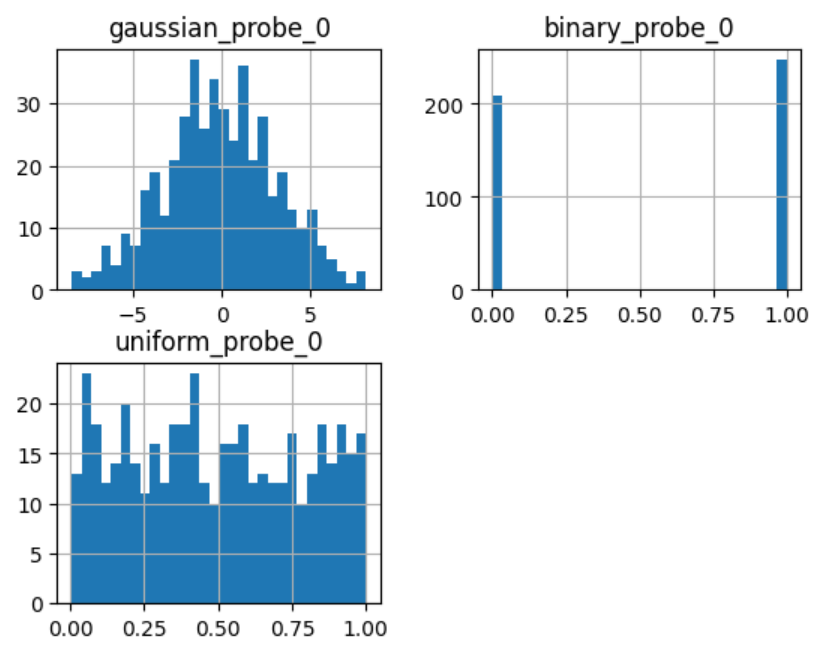
Let’s display the random features that the transformer created:
sel.probe_features_.head()
Here we find some example values of the probe features:
gaussian_probe_0 binary_probe_0 uniform_probe_0
0 -0.694150 1 0.983610
1 1.171840 1 0.765628
2 1.074892 1 0.991439
3 1.698733 0 0.668574
4 0.498702 0 0.192840
Let’s go ahead and plot histograms:
sel.probe_features_.hist(bins=30)
In the histograms we recognise the 3 well defined distributions:
Let’s display the importance of the random features
sel.feature_importances_.tail()
worst symmetry 0.009176
worst fractal dimension 0.007825
gaussian_probe_0 0.003765
binary_probe_0 0.000354
uniform_probe_0 0.002377
dtype: float64
We see that the binary feature has an extremely low importance, hence, when we take the average, the value is so small, that no feature will be dropped:
sel.features_to_drop_
The previous command returns and empty list:
[]
It is important to select a suitable probe feature distribution when trying to remove variables. If most variables are continuous, introduce features with normal and uniform distributions. If you have one hot encoded features or sparse matrices, binary features might be a better option.
Additional resources#
More info about this method can be found in these resources:
Kaggle Tips for Feature Engineering and Selection, by Gilberto Titericz.
Feature Selection: Beyond feature importance?, KDDNuggets.
For more details about this and other feature selection methods check out these resources:
Feature Selection for Machine Learning#
Or read our book:
Feature Selection in Machine Learning#
Both our book and course are suitable for beginners and more advanced data scientists alike. By purchasing them you are supporting Sole, the main developer of Feature-engine.
